Working with SQLite Database in Julia
Let's learn how to read data from the most popular database in the world
The SQLite library implements an embedded, relational and serverless database engine. This means that the database itself is simply a disk file with read-and-write access. The file format has cross-platform compatibility, and has therefore been deployed on millions of devices across the world. Due to its massive adoption across a wide variety of domains, data is often stored and shared as an SQLite database file (.sqlite extension). The SQLite project provides a command-line tool sqlite3, which is capable of performing all sorts of SQL queries. In case you want to learn more, I have written another blog post about it.
This post aims to explore SQLite support in Julia. It is provided via the SQLite.jl package, which is part of the general registry. To add it to your environment, simply execute ] add SQLite. We will also be making use of DataFrames.jl , which is another popular data analysis package.
Pluto notebook
All the code shown below has been executed in a Pluto notebook, which can be downloaded from my GitHub repository.
Reading the database
I am making use of a sample dataset obtained from Kaggle. It contains Neural Information Processing System (NIPS) papers, authors, text etc.
db = SQLite.DB("input_sqlite_database/nips_papers.sqlite")
Check schema
SQLite.tables(db)
3-element Vector{SQLite.DBTable}:
SQLite.DBTable("papers", Tables.Schema:
:id Union{Missing, Int64}
:year Union{Missing, Int64}
:title Union{Missing, String}
:event_type Union{Missing, String}
:pdf_name Union{Missing, String}
:abstract Union{Missing, String}
:paper_text Union{Missing, String})
SQLite.DBTable("authors", Tables.Schema:
:id Union{Missing, Int64}
:name Union{Missing, String})
SQLite.DBTable("paper_authors", Tables.Schema:
:id Union{Missing, Int64}
:paper_id Union{Missing, Int64}
:author_id Union{Missing, Int64})
As shown above, the database contains three tables - papers, authors and paper_authors. Individual columns of a table can be checked as follows:
SQLite.columns(db, "papers")
Convert to DataFrame
It is often useful to read the information from a SQLite database into a DataFrame object. This can be done as shown below:
df_papers = DBInterface.execute(db, "SELECT id, year, title FROM papers") |> DataFrame
Here we have selected only the id, year and title columns from the papers table. The resulting DataFrame should look like this:
Similarly, we can also read the authors table into a DataFrame, and then convert it into a Dict.
df_authors = DBInterface.execute(db, "SELECT id, name FROM authors") |> DataFrame
authors_dict = Pair.(df_authors.id, df_authors.name) |> Dict
Once more, we do the same for paper_authors table:
df_paper_authors = DBInterface.execute(db, "SELECT paper_id, author_id FROM paper_authors") |> DataFrame
Analyze data
Find all authors for a given year
To determine author names for all the years, we need to combine information from separate tables (DataFrames in our case). df_paper_authors has the mapping between author_id and paper_id. The author_id itself uniquely maps to an author name in authors_dict. Let's find out who wrote these papers.
function get_authors_all_years(authors_dict::Dict,
df_papers::DataFrame = df_papers,
df_paper_authors::DataFrame = df_paper_authors)
df_result = deepcopy(df_papers)
author_ids = Vector{Int64}[]
for p_id in df_result[!, :id]
df_filter = filter(row -> row.paper_id == p_id,
df_paper_authors)
push!(author_ids, df_filter[!, :author_id])
end
author_names = Vector{String}[]
for author_group in author_ids
names_group = String[]
for a_id in author_group
# Each id maps to a unique author name
push!(names_group, authors_dict[a_id])
end
push!(author_names, names_group)
end
# Merge author names into one row
authors = String[]
for author in author_names
push!(authors, join(author, ", "))
end
insertcols!(df_result, 3, :authors => authors)
return df_result
end
Executing the above function, we get:
Count all papers and group them by years
function count_papers(year_start::Int64,
year_end::Int64,
df_papers::DataFrame = df_papers)
gdf_papers = groupby(df_papers, :year)
df_count = combine(x -> length(x.title), gdf_papers)
return filter(row -> year_start ≤ row.year ≤ year_end, df_count)
end
Executing the above function, we get:
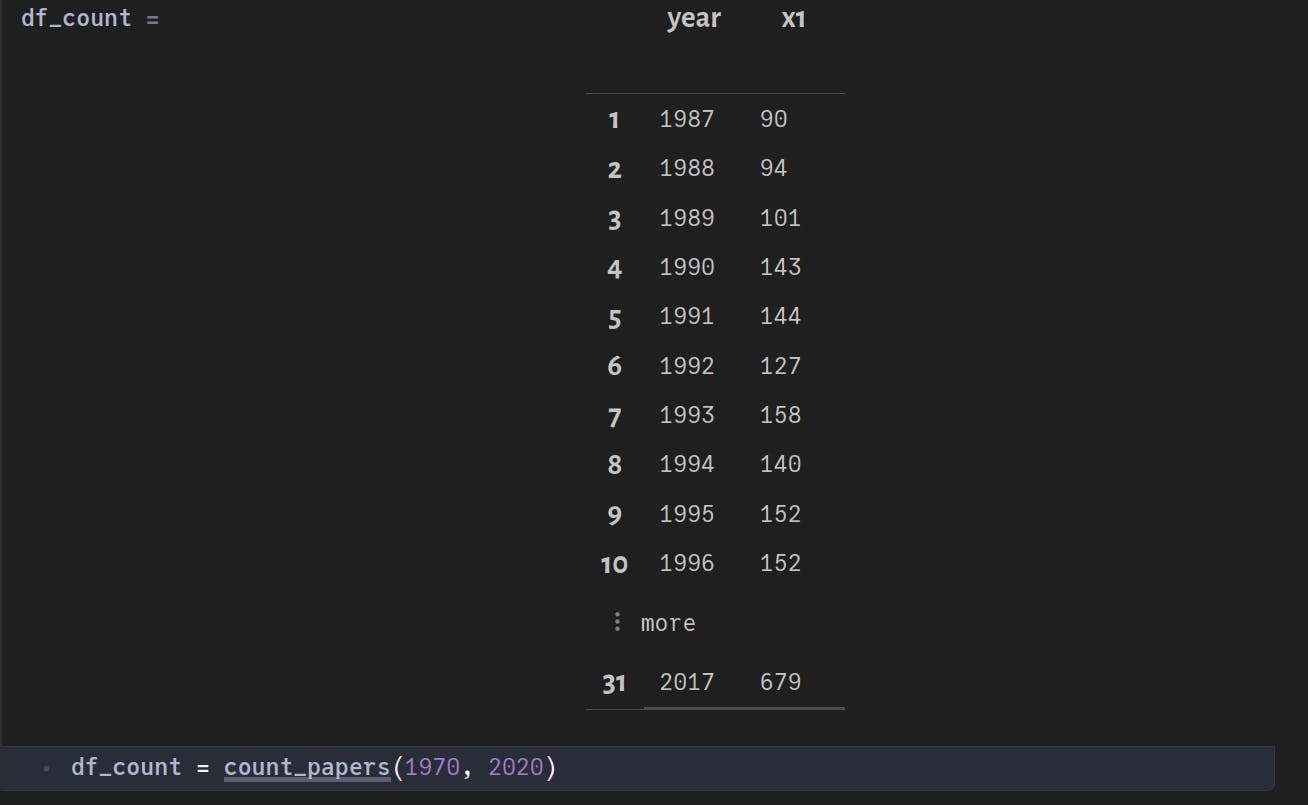
Save to database
Now that we have done some analysis, we can save our results to either the same SQLite database (as a new table) or to an entirely new one. The below function saves the df_input DataFrame to the SQLite database with the name db_name (creates new if it doesn't exist) in a table called table_name.
function save_to_db(df_input::DataFrame,
db_name::String,
table_name::String)
db_save = SQLite.DB(db_name)
SQLite.load!(df_input,
db_save,
table_name;
temp = false,
ifnotexists = false,
replace = false,
on_conflict = nothing,
analyze = false)
end
Let's try to save the df_all_authors and df_count DataFrames into two separate tables in a new database called papers_from_julia.sqlite:
save_to_db(df_all_authors, "output/papers_from_julia.sqlite", "papers")
save_to_db(df_count, "output/papers_from_julia.sqlite", "paper_count")
We can now verify the schema using the sqlite3 command-line tool:
sqlite> .open "output/papers_from_julia.sqlite"
sqlite> .databases
main: /home/vikas/Desktop/Julia_training/SQL_data_analysis/output/papers_from_julia.sqlite r/w
sqlite> .schema
CREATE TABLE IF NOT EXISTS "papers" ("id" INT NOT NULL,"year" INT NOT NULL,"authors" TEXT NOT NULL,"title" TEXT NOT NULL);
CREATE TABLE IF NOT EXISTS "paper_count" ("year" INT NOT NULL,"x1" INT NOT NULL);
Both the tables are present as expected. Let's check the contents of the papers table:
sqlite> SELECT * FROM papers
...> LIMIT 10;
+----+------+---------------------------------------+----------------------------------------------------------------------------------------------------+
| id | year | authors | title |
+----+------+---------------------------------------+----------------------------------------------------------------------------------------------------+
| 1 | 1987 | Hisashi Suzuki, Suguru Arimoto | Self-Organization of Associative Database and Its Applications |
| 2 | 1987 | Philip A. Chou | The Capacity of the Kanerva Associative Memory is Exponential |
| 3 | 1987 | Eric B. Baum, Frank Wilczek | Supervised Learning of Probability Distributions by Neural Networks |
| 4 | 1987 | John C. Platt, Alan H. Barr | Constrained Differential Optimization |
| 5 | 1987 | Ralph Linsker | Towards an Organizing Principle for a Layered Perceptual Network |
| 6 | 1987 | Gene A. Tagliarini, Edward W. Page | A Neural-Network Solution to the Concentrator Assignment Problem |
| 7 | 1987 | Ken Hsu, David Brady, Demetri Psaltis | Experimental Demonstrations of Optical Neural Computers |
| 8 | 1987 | Frank H. Eeckman | The Sigmoid Nonlinearity in Prepyriform Cortex |
| 9 | 1987 | Amir F. Atiya | Learning on a General Network |
| 10 | 1987 | Christopher L. Scofield | A Mean Field Theory of Layer IV of Visual Cortex and Its Application to Artificial Neural Networks |
+----+------+---------------------------------------+----------------------------------------------------------------------------------------------------+
All looks good. The author names are now present in a separate column.
Conclusion
The ability to read/write SQLite database files from Julia is extremely powerful for any data science workflow. Additionally, they can be easily parsed into a DataFrame type object. DataFrames themselves offer a wide array of analysis tools along with easy interoperability with numerous data visualization packages in the Julia ecosystem.
I hope you learned something interesting today. Don't forget to share this post within your network. Thank you for your time!
